5.4 GENETIC REGULATION OF 寿命 IN S. CEREVISIAE
The single-cell eukaryotic organism Saccharomyces cerevisiae has been used for many years in aging research. It is a model organism for this research because it is short-lived, is inexpensive to grow and maintain, and has well-characterized genetics and physiology. Commonly known as brewer‘s or bread yeast, S. cerevisiae was the first eukaryotic organism to have its genome sequenced (completed in 1996). The 12,156,590 base pairs in the DNA sequence are contained in a haploid set of 16 chromosomes. There are approximately 6600 known genes with an average size of 490 codons (1.47 kbp). The genome of this yeast is compact, with 75% of the DNA sequence as exons. Although researchers have identified many yeast strains having mutations that influence longevity, only a few of these have been screened in sufficient detail to warrant discussion here. Genetic mechanisms related to longevity in S. cerevisiae include genomic instability, gene silencing, and mutations in nutrient-responsive pathways. We take a look at each of these in more detail in this section, but first we need to briefly discuss how the yeast reproduces, because its reproductive cycle affects its longevity.
5.4.1 S. cerevisiae reproduces both asexually and sexually
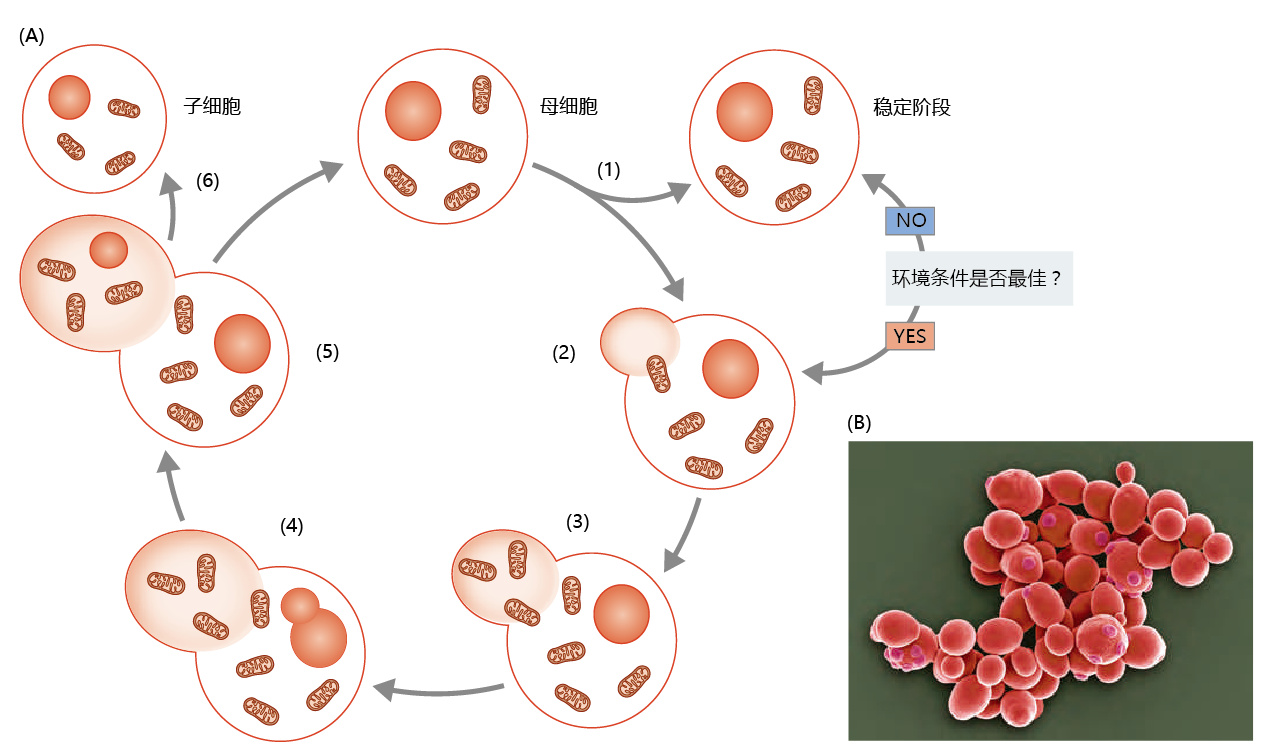
Yeast can exist in two chromosomal states, one diploid (containing two sets of chromosomes) and one haploid (containing only one set of chromosomes). The haploid cells reproduce mitotically, creating a new cell from the parent cell by budding. During budding, the nucleus of the mother cell undergoes mitosis to form a daughter nucleus, which migrates into the forming bud (Figure 5.20A). Chitin, a tough polymer of nitrogen-containing sugars, forms between the bud and the mother cell. When mitosis is complete, the new daughter cell separates from the mother cell (Figure 5.20B). After separation, a circular chitin residue known as the bud scar remains permanently on the surface of the mother cell.
Figure 5.20 The asexual reproductive cycle of S. cerevisiae. (A) Asexual reproduction occurs by budding. (1) The mother cell can sense whether 环境al conditions are optimal for reproduction. If they are suboptimal, the mother cell may enter a stationary phase until conditions improve. (2) If 环境条件适合繁殖,a bud forms as an extension of the mother cell‘s cytoplasm. (3) New cytoplasmic organelles are synthesized and migrate into the forming bud. (4) Mitosis begins, the genetic material is duplicated, and a new nucleus splits from the mother nucleus. (5) The newly formed nucleus migrates into the bud. (6) Cytokinesis occurs, and mother and daughter cells separate. (B) 子代酵母菌细胞的扫描电镜,forming as buds on mother cells. The number of daughter cells produced by a mother cell, as indicated by the number of scars left by daughter cells after separation, provides a measure of reproductive senescence. Note the smaller size of the newly separated daughter cells. (B, courtesy of S. Gschmeissner/Getty Images.)
Diploid reproduction occurs as a matter of chance and is regulated, at least in part, by 环境al conditions. Budding results in two types of daughter cells, a and α. If the two opposite mating types meet, they can fuse and enter the diploid phase of the cell cycle. In 环境al conditions that are favorable for the survival of daughter cells, the diploid yeast reproduces by mitosis, or budding. If, however, 环境al conditions are such that the survival of daughter cells is at risk, the diploid yeast produces four gametes called ascospores, two with a and two with α chromosomes. When 环境al conditions are more favorable, the spores germinate and produce four haploid yeast cells.
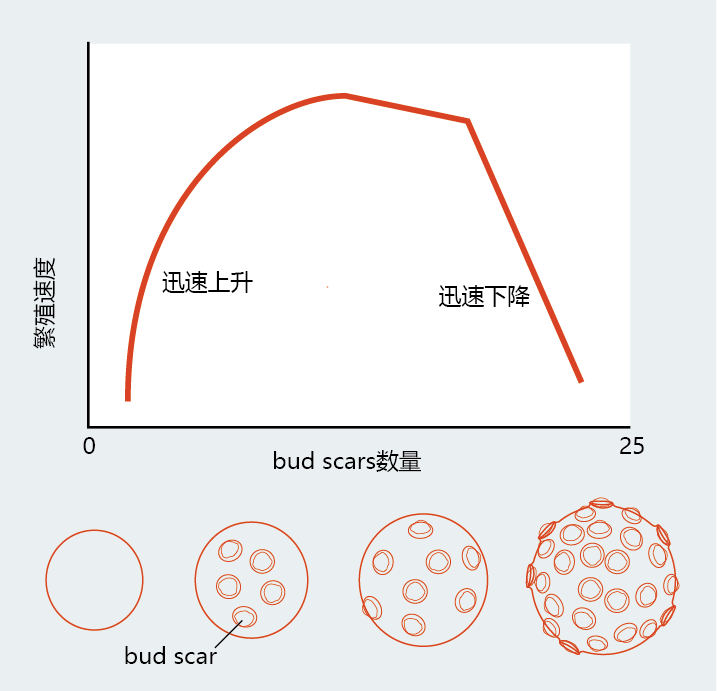
The life history of budding yeast can be described by the Gompertz mortality equation, which distinguishes replicative senescence in the single-cell eukaryotic yeast from mitotic cells derived from the tissue of multicellular organisms. As shown in Figure 5.21, the number of bud scars and the size of the mother cell provide convenient markers for estimating the reproductive age of the mother cell. Individual S. cerevisiae produce an average of 20–30 new daughter cells (20–30 bud scars) before entering replicative senescence. However, neither the number of bud scars nor the size of the mother cells is the direct cause of senescence. Mutations that increase the number of bud scars and/or cell size do not affect longevity.
Figure 5.21 Bud scars and replicative senescence of S. cerevisiae. A young yeast cell is small, has few bud scars, and undergoes a rapid rate of budding. As shown in the graph, the initial burst of reproduction is followed by an extended period with a stable reproductive rate. As the yeast approaches its maximum number of buddings, reproduction rapidly declines and the organism dies.
5.4.2 环境条件影响生殖和寿命
In Chapter 3, we showed that the proximal 环境 has a significant influence on the life history pattern and reproductive success in populations of single-cell organisms such as S. cerevisiae. The life history of S. cerevisiae shows the typical pattern associated with wild, single-cell populations: (1) a rapid increase in population when resources (food, space, etc.) are abundant; (2) stabilization of population size when resources match the intrinsic rate of growth (r = reproduction rate − death rate = 0); (3) decrease in population size (decrease in reproduction) as resources begin to decline; and (4) replicative senescence and death of the population when resources are exhausted.
S.cerevisiae has two reproductive strategies that help extend its reproductive life span and prevent the death of the population during times of limited or depleted 环境al resources. Sexual reproduction of the diploid yeast produces ascospores, which can survive several years and can withstand many different types of 环境al conditions. This provides a long-term solution for extending reproduction during times of 环境al stress. The budding of haploid yeast provides a short-term solution for extending the reproductive period. When a budding yeast detects that 环境al conditions are detrimental to the survival of a daughter cell, asexual reproduction (budding) is turned off. The budding yeast enters a nondividing state, called the stationary phase, but remains metabolically active. If the 环境 conditions improve within 1–2 weeks, the yeast resumes reproductive activity; otherwise, the cell dies.
In the stationary phase, the yeast has significantly elevated levels of oxygen radicals toward the end of its life span. In this phase, mutant yeast cells without genes for superoxide dismutase (SOD) have shorter life spans, whereas mutants that overexpress SOD have significantly longer post-reproductive life spans. The increase in oxygen radicals in the nondividing yeast seems to stimulate the p53 checkpoint protein, which, in turn, results in an apoptotic-like death. The nondividing yeast has senescence characteristics similar to those of more complex organisms, thus providing investigators with a simple eukaryotic organism in which to study the genetics of longevity.
5.4.3 Structural alteration in DNA affects life span
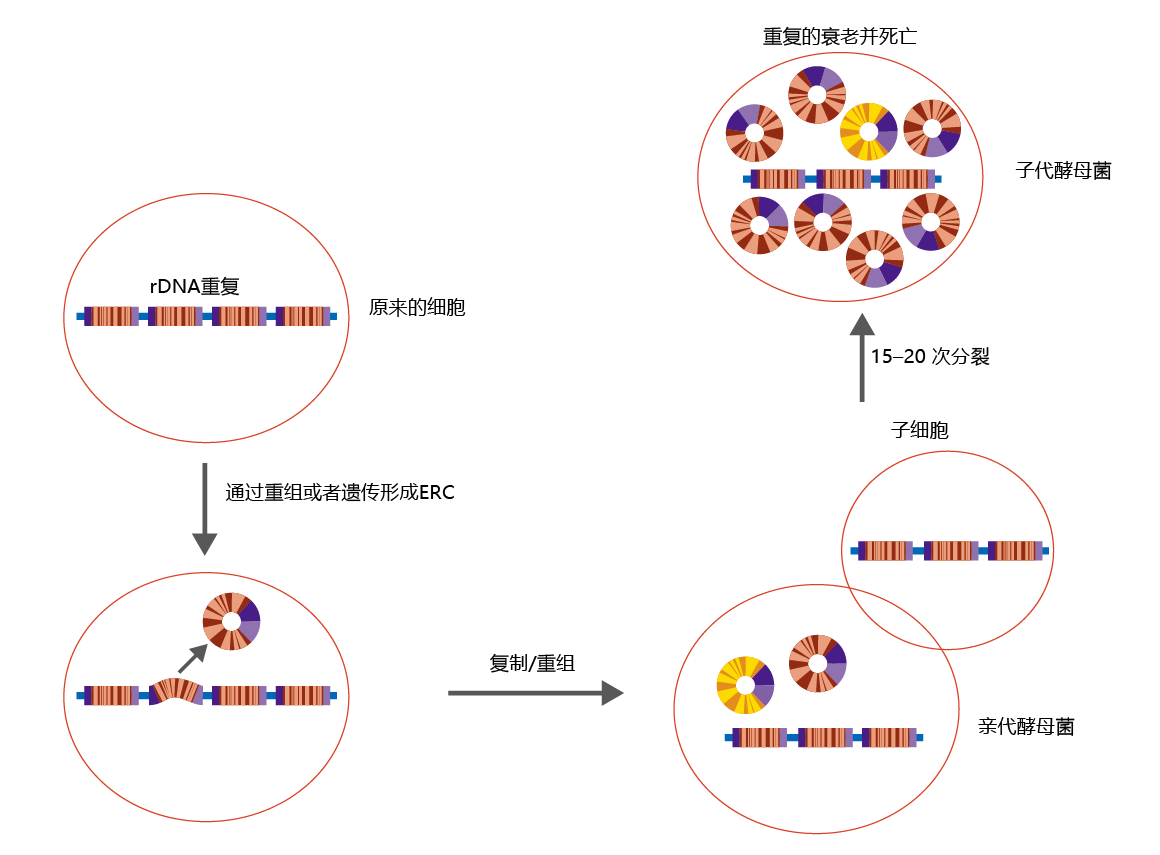
The genetic mechanisms that account for replicative senescence in the budding yeast remain, in large part, unknown. However, genomic instability, characterized by a structural alteration in the DNA, appears to play a fundamental role in the yeast life span. The genomic instability associated with aging yeast occurs in the ribosomal DNA (rDNA), located in the nucleolus. The yeast rDNA locus is a segment on Chromosome XII, containing 100–200 repeats of 9 kbp, arranged in tandem, that codes for rRNA. The repetitive nature of the rDNA makes these areas highly susceptible to intrachromosomal homologous recombination, also known as general recombination. Although general recombination is an essential part of meiosis, this process in yeast also gives rise to potentially damaging extrachromosomal rDNA circles (ERCs), as shown in Figure 5.22. The ERCs contain a replication origin site and thus can self-propagate, leading to an accumulation of these structures in the mother yeast. The accumulation of ERCs correlates highly with replicative senescence, although how ERCs cause replicative senescence has yet to be demonstrated. Studies in S. cerevisiae suggest that the production of ERCs and the subsequent replicative senescence may be related to the acetylation-deacetylation mechanism of gene expression regulation.
Figure 5.22 Formation of extrachromosomal rDNA circles (ERCs) and yeast replicative senescence. The budding yeast contains highly repetitive sequences of rDNA, occurring in tandem, that form ERCs through general recombination. ERCs form in the mother cell after each round of budding. The accumulation of ERCs is associated with replicative senescence and death of the yeast.
The ERCs are a “biological clock” of sorts, associated with the life span of S. cerevisiae. This conclusion is the result of two findings. First, budding yeast containing a mutation that prevents the formation of ERCs live significantly longer than wild-type yeast. Second, daughter cells that arise from old mother yeast cells receive ERCs and have significantly shorter life spans than ERC-free daughter yeast.
5.4.4 The SIR2 pathway is linked to 寿命
Silencing in the transcription of rDNA into rRNA appears to be regulated by a highly conserved gene named silent information regulator 2 (sir2). Overexpression of sir3 results in decreased rDNA expression and reduced formation of ERCs. Overexpression of sir2 doubles the yeast's replicative life span. The means by which the sir2 gene and its encoded 蛋白质 (SIR2) cause extension of the yeast replicative life span has not been completely worked out. We do know, however, that the SIR2 蛋白质 is a histone deacetylase, which causes compaction of the chromatin and gene silencing.
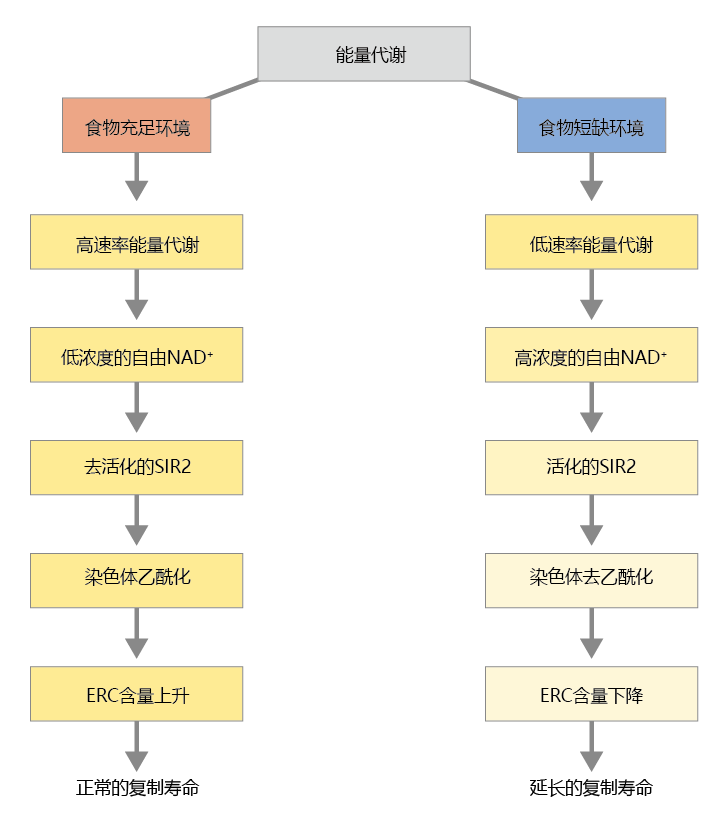
SIR2-induced gene silencing seems to be connected directly to the pathways that convert food into energy. As described in Chapter 4, electrons arising from the oxidation of glucose are shuttled to the ATP-synthesizing apparatus by electron carriers. One of these carriers, oxidized nicotinamide adenine dinucleotide (NAD+ ), activates or “turns on” the SIR2 蛋白质. Thus, the cell concentration of NAD+ directly affects the level of SIR2 activity; low concentrations of NAD+ deactivate the SIR2 蛋白质; high concentrations of NAD+ activate the SIR2 protein. The NAD+ concentration fluctuates with the amount of food in the yeast‘s 环境. Times of plenty lead to increased reproduction and relatively high rates of metabolic activity. In turn, the metabolic pathways generate increased electron flow and a lower ratio of NAD+ to NADH+ H+ (Figure 5.23) . Thus, the activity of the SIR2 蛋白质 declines, rDNA is transcribed, and reproduction (budding) proceeds. Conversely, when food becomes scarce, metabolism decreases and the resulting diminished electron flow increases the ratio of NAD+ to NADH+ H+ . In turn, NAD+ binds to SIR2, causing activation of the histone deacetylase and compaction of the chromatin, silencing of the rDNA locus, reduction of the budding process, and an extended replicative life span for the yeast.
Figure 5.23 A proposed mechanism that links energy metabolism in S. cerevisiae to an extended life span. Times of plenty lead to increased reproduction and relatively high rates of metabolic activity (left). During periods of scarce food supply and low reproduction, energy metabolism in the yeast is reduced (right), leading to decreased rDNA transcription, reduced budding, and inhibition of ERC formation.
5.4.5 Loss-of-function mutations in nutrient-responsive pathways may extend the life span
The amount of food in the yeast‘s 环境 controls, in large part, the rate of reproduction and thus the length of life. An abundance of food results in high rates of reproduction and shorter life span. Food shortages delay reproduction and extend life span. The way in which the yeast recognizes the amount of food in its 环境 and then alters reproduction appears to be related to specific nutrients.
The biochemical or physiological link among extracellular nutrients, reproduction, and life span may involve highly conserved cellular signaling pathways. These pathways in yeast begin when a receptor on the cell membrane binds nutrients or nutrient metabolites. At least one of the signaling pathways responds to the uptake of glucose and may involve the regulation of sir2, but the details of this pathway are not clear. Other pathways are initiated through ligands, molecules that bind specifically to receptor sites on other molecules. Certain ligands bind to G-蛋白质-coupled receptors, a type of receptor known to initiate signaling cascades linking nutrient availability to the expression of genes important to growth and reproduction in all eukaryotes.
Two general nutrient-responsive pathways have been identified that alter life span in yeast: target of rapamycin (TOR) and 蛋白质 kinase A (PKA). Mutations in these two pathways extend life span by inhibiting enzymes that catalyze the phosphorylation of 蛋白质, although each pathway is independent. While the details of how TOR and PKA extend longevity are unclear, the mechanisms undoubtedly involve components of the highly conserved G-蛋白质 signaling process.